Designing Transposable Elements-Specific sgRNA
Introduction
CRISPR-TE is a tool that uses the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology to design single-guide RNAs (sgRNAs) that can be used to target transposable elements (TEs) in the genome. CRISPR-TE provides two strategies for designing sgRNAs that target TEs: TE single duplicates and TE subfamilies.
In the TE single duplicate mode, CRISPR-TE generates all possible sgRNAs for a specified TE, along with detailed information such as coordinates, active score, mismatch count, and annotation. This allows users to select the sgRNAs with the highest targeting accuracy and the lowest off-target effects.
In the TE subfamily mode, CRISPR-TE uses a combination of sgRNAs to target multiple TEs within a subfamily. This allows for more efficient and effective targeting of a group of related TEs. The tool uses a greedy algorithm to find the optimal sgRNA combinations for editing TE subfamilies, taking into account off-target effects.
Overall, CRISPR-TE is a useful tool for researchers studying transposable elements and looking for ways to target and manipulate them in the genome. By using CRISPR technology, it enables precise and efficient editing of TEs, which can help advance our understanding of their role in genomic instability and other genetic phenomena.
CRISPR-TE Input Section
To use CRISPR-TE, users need to specify three pieces of information:
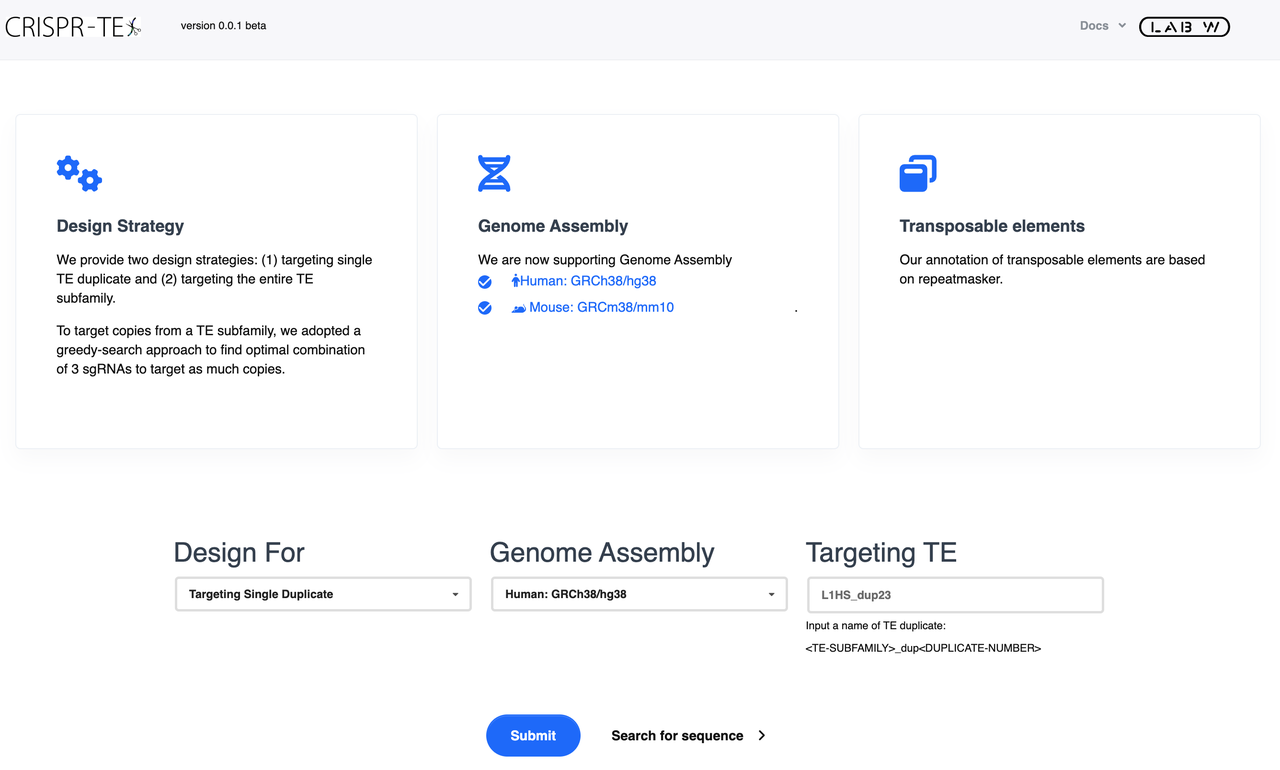
-
Design purpose: Select your purpose for TE editing, at single duplicate level or subfamily level. You can scroll through and select from the drop-down.
-
Genome Assembly:CRISPR-TE provides human(hg38) and mouse(mm10) species. Select your genome of interest from the list. You can scroll through and select from the drop-down or type and search the names of the species
-
Targeting TE: input which TE you want to target. For example, L1HS_dup23 at single duplicate level and SVA_D at subfamily level.
CRISPR-TE Output section
CRISPR-TE generates two main output depending on the user's intended purpose.
TE single duplicate level
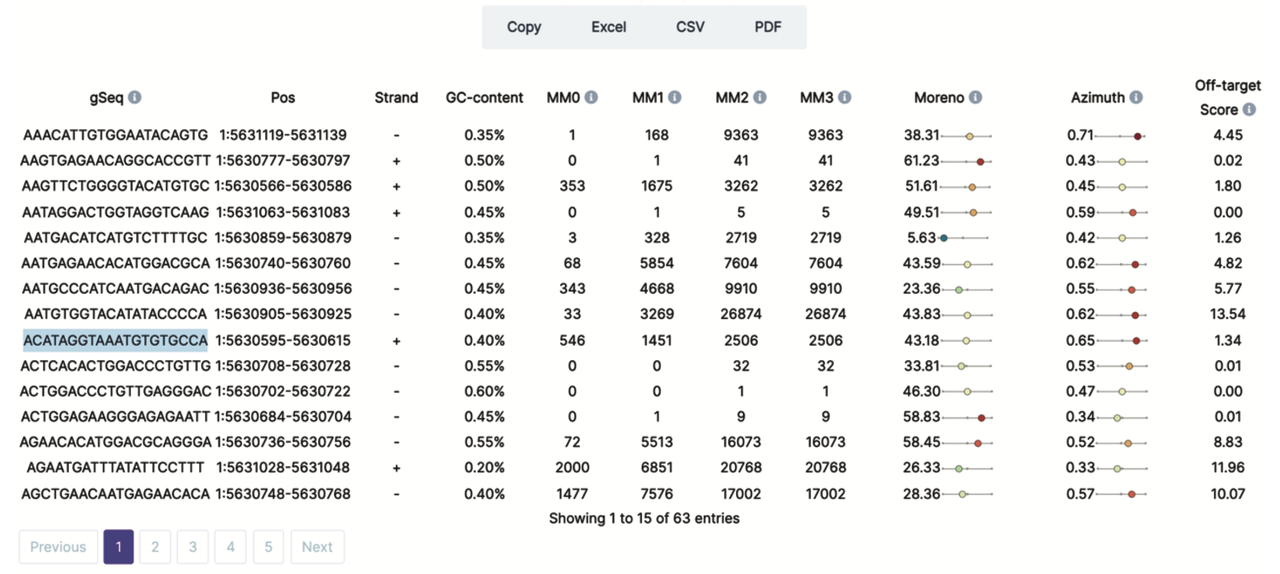
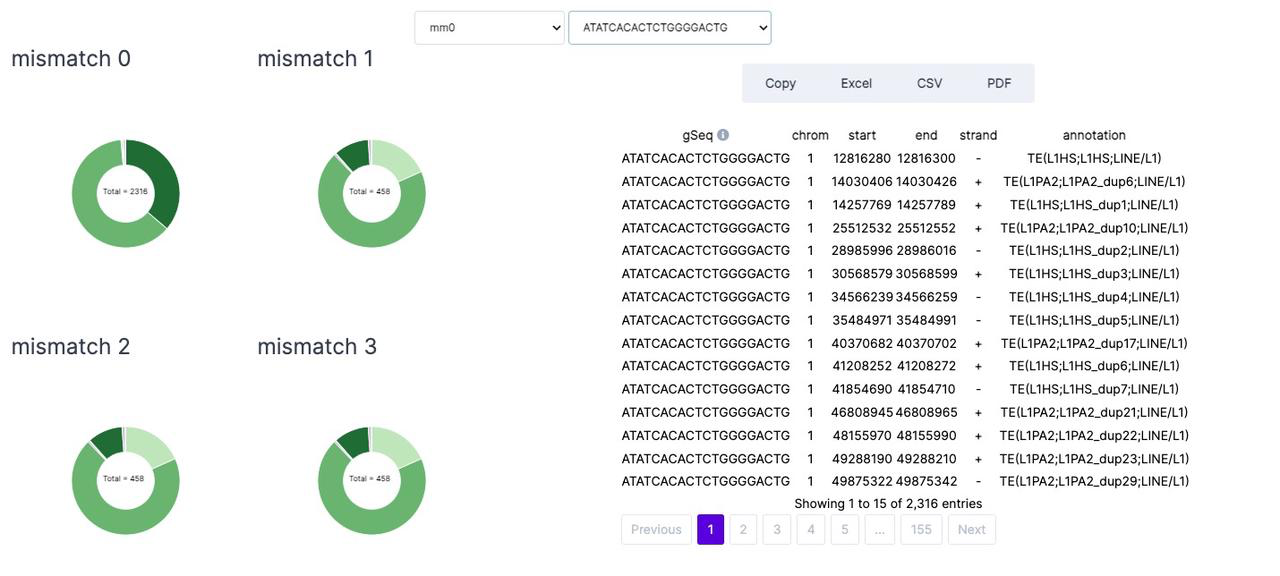
-
gSeq: sgRNA sequence
-
Pos: sgRNA coordinates in the genome
-
Strand: sgRNA strand
-
GC-content: the percentage of nitrogenous bases in a DNA
-
MM0: numbers of perfect target site in the genome
-
MM1,2,3: potential off-targets number with 1, 2, 3 mismatches, mismatch is defined by the he Hamming distance considered between the guide RNAs and corresponding off-target sequences
-
Moreno, Azimuth: sgRNA on-target activity score calculated using the algorithm in Moreno-Mateos, M. A. et al., Nature Methods 2015 and Mendoza, B. J. & Trinh, C. T, Bioinforamtics 2018.
-
Off-target score: sgRNA off-target score calculated using the customized formula below:
- According to the annotation of sgRNA target site, we give different penalties. If sgRNA off-target to diffrent TE subfamily, it will have the hightest penalty. The priority order of other genomic class is promoter_TSS, exon, intron and intergenic.
- Score = on_target_percentage - off_target_te * 0.001 - (0.04 * off_target_on_promoter_TSS + 0.03 * off_target_on_exon + 0.02 * off_target_on_intron + 0.01 * off_target_on_intergenic) * 0.0001
The main ouput of CRISPR-TE is a page that displays the detailed information of possible target sgRNA given target TE single duplicate. These information including:
TE subfamily level
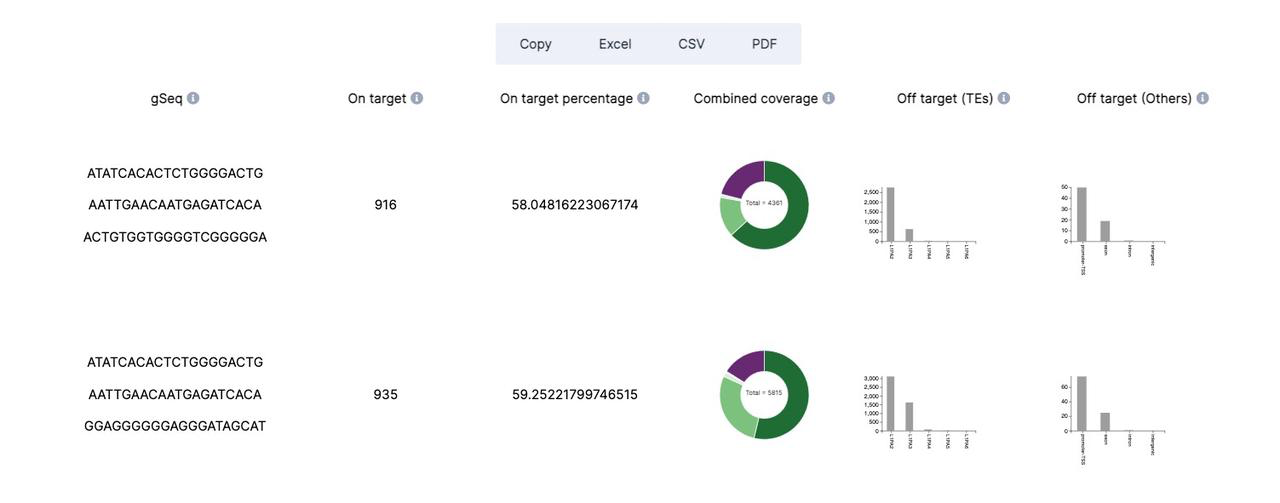
- gSeq combination: gRNA combinations for targeting TE subfamily
- On-target, on-target percentage: the sgRNA combination on-target number and percentage
- Combined coverage: the pie chart of sgRNA combination target result, detailed target TE information will display when mouse is hover.
- Off-target(TEs/Others): The top5 off-target transposable elements and other genomic class of sgRNA combination.
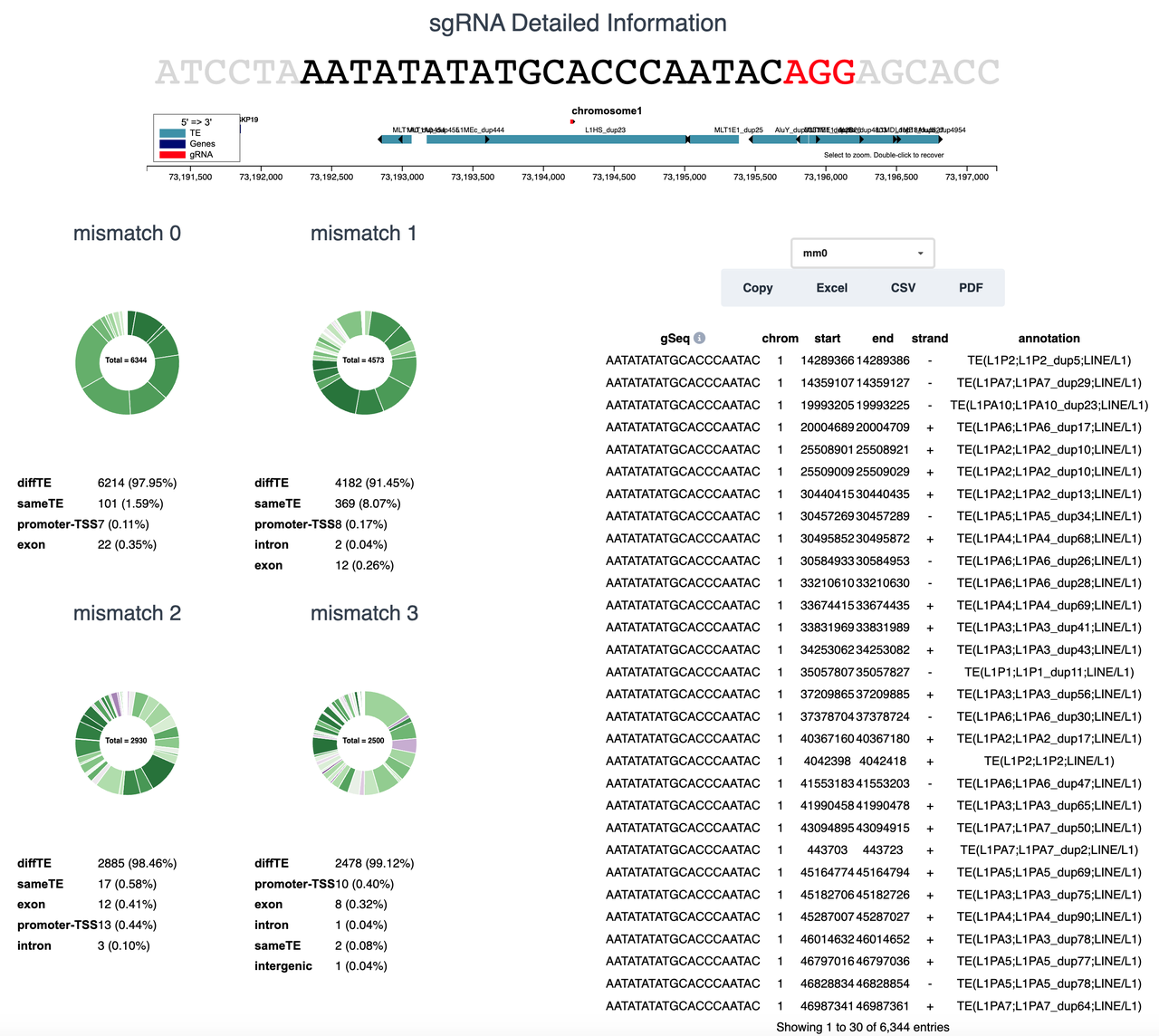
- Target summary pie chart of individual sgRNA with mismatch 0, 1, 2, 3
- Target site with coordinates and detailed annotation of each sgRNA from user selected combinations. Users can select up to three mismatches in the drop-down.
The other ouput of CRISPR-TE is a page that shows the detailed information of sgRNA combinations given target TE subfamily.
CRISPR-TE also provides detailed information of each individual sgRNA from sgRNA combinations.
Scoring
Initial score is calcualted according to Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12, 982–988 (2015) and Doench et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9 (2016).Feature (the initial sgRNA-score is calculated by adding up of the values of all features found in the sequence at position i)
| Pos | A | T | C | G | AA | AT | AC | AG | TA | TT | TC | TG | CA | CT | CC | CG | GA | GT | GC | GG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -6 | 0 | 0 | 0 | -0.0297 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| -5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| -4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0229 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0247 | 0 |
| -3 | -0.0422 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| -2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| -1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0.0024 | 0.1146 | 0.0000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0542 | -0.0016 |
| 4 | 0 | 0 | 0 | 0.0677 | -0.0169 | 0 | 0 | 0.0637 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0268 |
| 5 | -0.0183 | 0 | 0 | 0.0209 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0537 | 0 | 0 |
| 6 | 0.0116 | 0 | 0 | 0.0276 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0419 |
| 7 | 0.0176 | 0.0354 | 0.0695 | 0.0614 | 0 | 0 | 0 | 0 | 0 | -0.0862 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0744 |
| 8 | -0.0185 | 0 | 0 | 0.0251 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | -0.0106 | 0 | -0.0005 | 0.0511 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0534 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | -0.0216 | 0.0380 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0.0155 | 0 | 0 | 0 | 0 | 0 | 0.0349 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | -0.0338 | 0 | 0 | 0.0165 | 0 | 0 | 0 | 0 | 0 | -0.0944 | 0 | 0.0459 | 0 | 0 | 0 | 0 | 0 | 0.0024 | 0 | 0 |
| 13 | -0.0156 | 0 | 0 | 0.0179 | -0.0974 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0000 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0.0151 | 0.0340 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0097 | 0 | 0 | 0 |
| 15 | 0 | -0.0687 | 0.0343 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0889 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0.0319 | 0.0132 | 0.0376 | 0.1011 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | -0.0014 | 0 | 0 | 0 | 0 | 0 | -0.0543 | 0 | 0 | 0 | -0.0664 | 0 | 0 | 0.0951 | 0 | 0 | 0.1067 | 0 | 0 |
| 18 | 0.0375 | -0.0120 | 0.0530 | 0.1055 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0622 | 0 | 0 | 0 | 0 | 0.0610 | 0 | 0 |
| 19 | 0.0105 | 0 | -0.0316 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | -0.0735 | 0.1116 |
| 20 | 0 | 0 | -0.0004 | 0.0361 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | -0.0843 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0.0191 | -0.0003 | 0.0799 | 0.0852 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | PAM | |||||||||||||||||||
| 23 | PAM | |||||||||||||||||||
| 24 | 0 | 0.0173 | -0.0132 | -0.0463 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0578 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | 0.0016 | 0 | -0.0307 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0481 | 0 | 0 | 0 | 0 | -0.0123 | 0.0734 | 0 | 0 |
| 26 | 0.0124 | 0 | 0 | 0.0307 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 28 | 0 | -0.0176 | 0 | -0.0142 | 0 | 0 | 0 | -0.0419 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | -0.0378 | 0 | 0 | 0 |
| 29 | 0.0486 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |